# install the package
install.packages("tidyverse")Manipulating, analyzing, and exporting data with tidyverse
Data manipulation using dpylr and tidyr
We’ve learned to subset with brackets [], which is useful, but can become cumbersome with complex subsetting operations. Enter the two packages below, both of which are part of the tidyverse.
dplyr– a package that is useful for tabular data manipulationtidyr– a package that is useful for donverting between data formats used in plotting / analysis
The tidyverse package tries to address 3 common issues that arise when doing data analysis in R:
- The results from a base
Rfunction sometimes depend on the type of data. Rexpressions are used in a non standard way, which can be confusing for new learners.- The existence of hidden arguments having default operations that new learners are not aware of.
You should have tidyverse installed already. If you do not, you can type the following into the console.
# load the package
library("tidyverse")── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
✔ ggplot2 3.4.0 ✔ purrr 0.3.5
✔ tibble 3.1.8 ✔ dplyr 1.0.10
✔ tidyr 1.2.1 ✔ stringr 1.4.1
✔ readr 2.1.3 ✔ forcats 0.5.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()Let’s read in our data using read_csv(), from the tidyverse package, readr.
surveys <- read_csv("~/data-carpentry/data_raw/portal_data_joined.csv") #read in dataRows: 34786 Columns: 13
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (6): species_id, sex, genus, species, taxa, plot_type
dbl (7): record_id, month, day, year, plot_id, hindfoot_length, weight
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.str(surveys) #inspect the data
view(surveys) #preview the dataNext, we’re going to learn some of the most common dplyr functions
select(): subset columnsfilter(): subset rows on conditionsmutate(): create new columns by using information from other columnsgroup_by()andsummarize(): create summary statistics on grouped dataarrange(): sort resultscount(): count discrete values
Selecting columns and filtering rows
To select columns of a dataframe, we use select().
select(datafame, columns_to_keep)
select(surveys, plot_id, species_id, weight)# A tibble: 34,786 × 3
plot_id species_id weight
<dbl> <chr> <dbl>
1 2 NL NA
2 2 NL NA
3 2 NL NA
4 2 NL NA
5 2 NL NA
6 2 NL NA
7 2 NL NA
8 2 NL NA
9 2 NL 218
10 2 NL NA
# … with 34,776 more rowsTo EXCLUDE a specific column, put - in front of the variable.
select(surveys, -record_id, -species_id)# A tibble: 34,786 × 11
month day year plot_id sex hindfoot…¹ weight genus species taxa plot_…²
<dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr>
1 7 16 1977 2 M 32 NA Neot… albigu… Rode… Control
2 8 19 1977 2 M 31 NA Neot… albigu… Rode… Control
3 9 13 1977 2 <NA> NA NA Neot… albigu… Rode… Control
4 10 16 1977 2 <NA> NA NA Neot… albigu… Rode… Control
5 11 12 1977 2 <NA> NA NA Neot… albigu… Rode… Control
6 11 12 1977 2 <NA> NA NA Neot… albigu… Rode… Control
7 12 10 1977 2 <NA> NA NA Neot… albigu… Rode… Control
8 1 8 1978 2 <NA> NA NA Neot… albigu… Rode… Control
9 2 18 1978 2 M NA 218 Neot… albigu… Rode… Control
10 3 11 1978 2 <NA> NA NA Neot… albigu… Rode… Control
# … with 34,776 more rows, and abbreviated variable names ¹hindfoot_length,
# ²plot_typeTo choose specific rows based on criteria, we use filter()
filter(surveys, year == 1995)# A tibble: 1,180 × 13
record…¹ month day year plot_id speci…² sex hindf…³ weight genus species
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr> <chr>
1 22314 6 7 1995 2 NL M 34 NA Neot… albigu…
2 22728 9 23 1995 2 NL F 32 165 Neot… albigu…
3 22899 10 28 1995 2 NL F 32 171 Neot… albigu…
4 23032 12 2 1995 2 NL F 33 NA Neot… albigu…
5 22003 1 11 1995 2 DM M 37 41 Dipo… merria…
6 22042 2 4 1995 2 DM F 36 45 Dipo… merria…
7 22044 2 4 1995 2 DM M 37 46 Dipo… merria…
8 22105 3 4 1995 2 DM F 37 49 Dipo… merria…
9 22109 3 4 1995 2 DM M 37 46 Dipo… merria…
10 22168 4 1 1995 2 DM M 36 48 Dipo… merria…
# … with 1,170 more rows, 2 more variables: taxa <chr>, plot_type <chr>, and
# abbreviated variable names ¹record_id, ²species_id, ³hindfoot_lengthConnecting inputs and outputs with pipes
We often want to perform multiple manipulations to our datasets. This can be done in a few ways: intermediate steps, nested functions, or pipes.
With intermediate steps, a temporary dataframe is created and used as input into the next function.
surveys2 <- filter(surveys, weight < 5) #filer surveys to create surveys2, the intermediate file
surveys_sml <- select(surveys2, species_id, sex, weight) #use surveys2 as input for the next step While sometimes useful (for initial validation, etc.), this can clutter the workspace and make it difficult to follow.
Nested functions fit one function inside of another.
surveys_sml <- select(filter(surveys, weight < 5), species_id, sex, weight) # filtering first, then selectingThis is handy, but can be difficult to read if too many functions are nested, as R evaluates the expression from the inside out (in this case, filtering, then selecting).
Pipes (%>%) are a recent addition to R and let you take the output of one function and send it directly to the next function as input. The package enabling this is magrittr, installed when you install dplyr.
surveys %>% # send surveys to filter
filter(weight < 5) %>% # use surveys as input, filter and send output to select
select(species_id, sex, weight) # use filtered data as input, select, and send output to console# A tibble: 17 × 3
species_id sex weight
<chr> <chr> <dbl>
1 PF F 4
2 PF F 4
3 PF M 4
4 RM F 4
5 RM M 4
6 PF <NA> 4
7 PP M 4
8 RM M 4
9 RM M 4
10 RM M 4
11 PF M 4
12 PF F 4
13 RM M 4
14 RM M 4
15 RM F 4
16 RM M 4
17 RM M 4Let’s break it down a bit.
- We use the pipe to send the
surveysdataset throughfilter() - Filter takes that input, and performs the filtering to keep rows where
weightis less than 5, then send the output to select - Select takes that input, and keeps only the
species_id,sex, andweightcolumns.
Note that since %>% takesthe object on its left and passes it as the first argument to the function on its right, we don’t need to explicitly include the data frame as an argument to the filter() and select() functions any more.
Some may find it helpful to read the pipe like the word “then.” For instance, in the example above, we took the dataframe surveys, then we filtered for rows with weight < 5, then we selected columns species_id, sex, and weight. The dplyr functions by themselves are somewhat simple, but by combining them into linear workflows with the pipe we can accomplish more complex manipulations of dataframes.
If we want to create a new object with this smaller version of the data, we can assign it a new name using the assignment operator.
surveys_sml <- surveys %>%
filter(weight < 5) %>%
select(species_id, sex, weight)
surveys_smlNote that the final dataframe is the leftmost part of this expression.
Challenge
Q&A: Using pipes, subset the surveys data to include animals collected before 1995 and retain only the columns year, sex, and weight.
Mutate
Frequently you’ll want to create new columns based on the values in existing columns, for example to do unit conversions, or to find the ratio of values in two columns. For this we’ll use mutate().
To create a new column of weight in kg
surveys %>%
mutate(weight_kg = weight / 1000) #assign the column name to the left of the "="# A tibble: 34,786 × 14
record…¹ month day year plot_id speci…² sex hindf…³ weight genus species
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr> <chr>
1 1 7 16 1977 2 NL M 32 NA Neot… albigu…
2 72 8 19 1977 2 NL M 31 NA Neot… albigu…
3 224 9 13 1977 2 NL <NA> NA NA Neot… albigu…
4 266 10 16 1977 2 NL <NA> NA NA Neot… albigu…
5 349 11 12 1977 2 NL <NA> NA NA Neot… albigu…
6 363 11 12 1977 2 NL <NA> NA NA Neot… albigu…
7 435 12 10 1977 2 NL <NA> NA NA Neot… albigu…
8 506 1 8 1978 2 NL <NA> NA NA Neot… albigu…
9 588 2 18 1978 2 NL M NA 218 Neot… albigu…
10 661 3 11 1978 2 NL <NA> NA NA Neot… albigu…
# … with 34,776 more rows, 3 more variables: taxa <chr>, plot_type <chr>,
# weight_kg <dbl>, and abbreviated variable names ¹record_id, ²species_id,
# ³hindfoot_lengthYou can also create a second new column based on the first new column within the same call of mutate()
surveys %>%
mutate(weight_kg = weight / 1000,
weight_lb = weight_kg * 2.2)# A tibble: 34,786 × 15
record…¹ month day year plot_id speci…² sex hindf…³ weight genus species
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr> <chr>
1 1 7 16 1977 2 NL M 32 NA Neot… albigu…
2 72 8 19 1977 2 NL M 31 NA Neot… albigu…
3 224 9 13 1977 2 NL <NA> NA NA Neot… albigu…
4 266 10 16 1977 2 NL <NA> NA NA Neot… albigu…
5 349 11 12 1977 2 NL <NA> NA NA Neot… albigu…
6 363 11 12 1977 2 NL <NA> NA NA Neot… albigu…
7 435 12 10 1977 2 NL <NA> NA NA Neot… albigu…
8 506 1 8 1978 2 NL <NA> NA NA Neot… albigu…
9 588 2 18 1978 2 NL M NA 218 Neot… albigu…
10 661 3 11 1978 2 NL <NA> NA NA Neot… albigu…
# … with 34,776 more rows, 4 more variables: taxa <chr>, plot_type <chr>,
# weight_kg <dbl>, weight_lb <dbl>, and abbreviated variable names
# ¹record_id, ²species_id, ³hindfoot_lengthIf this runs off your screen and you just want to see the first few rows, you can use a pipe to view the head() of the data. (Pipes work with non-dplyr functions, too, as long as the dplyr or magrittr package is loaded).
surveys %>%
mutate(weight_kg = weight / 1000) %>%
head()# A tibble: 6 × 14
record_id month day year plot_id speci…¹ sex hindf…² weight genus species
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr> <chr>
1 1 7 16 1977 2 NL M 32 NA Neot… albigu…
2 72 8 19 1977 2 NL M 31 NA Neot… albigu…
3 224 9 13 1977 2 NL <NA> NA NA Neot… albigu…
4 266 10 16 1977 2 NL <NA> NA NA Neot… albigu…
5 349 11 12 1977 2 NL <NA> NA NA Neot… albigu…
6 363 11 12 1977 2 NL <NA> NA NA Neot… albigu…
# … with 3 more variables: taxa <chr>, plot_type <chr>, weight_kg <dbl>, and
# abbreviated variable names ¹species_id, ²hindfoot_lengthThe first few rows of the output are full of NAs, so if we wanted to remove those we could insert a filter() in the chain, using the !is.na() approach we used in past sessions.
surveys %>%
filter(!is.na(weight)) %>%
mutate(weight_kg = weight / 1000) %>%
head()# A tibble: 6 × 14
record_id month day year plot_id speci…¹ sex hindf…² weight genus species
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr> <chr>
1 588 2 18 1978 2 NL M NA 218 Neot… albigu…
2 845 5 6 1978 2 NL M 32 204 Neot… albigu…
3 990 6 9 1978 2 NL M NA 200 Neot… albigu…
4 1164 8 5 1978 2 NL M 34 199 Neot… albigu…
5 1261 9 4 1978 2 NL M 32 197 Neot… albigu…
6 1453 11 5 1978 2 NL M NA 218 Neot… albigu…
# … with 3 more variables: taxa <chr>, plot_type <chr>, weight_kg <dbl>, and
# abbreviated variable names ¹species_id, ²hindfoot_lengthChallenge
Q&A: Create a new data frame from the surveys data that meets the following criteria:
Contains only the species_id column and a new column called hindfoot_cm containing the hindfoot_length values (currently in mm) converted to centimeters.
In this hindfoot_cm column, there are no NAs and all values are less than 3.
Hint: think about how the commands should be ordered to produce this data frame!
Split-apply-combine data analysis and the summarize() function
Many data analysis tasks can be approached using the split-apply-combine paradigm:
- split the data into groups
- apply some analysis to each group
- and then combine the results.
Key functions of dplyr for this workflow are group_by() and summarize()
The group_by() and summarize() functions
group_by() is often used together with summarize(), which collapses each group into a single-row summary of that group
group_by() takes as arguments the column names that contain the categorical variables for which you want to calculate the summary statistics.
So to compute the mean weight by sex
surveys %>%
group_by(sex) %>% # group by values categorical sex column
summarize(mean_weight = mean(weight, na.rm = TRUE)) # calculate the mean weight for each category in the sex column, save as a new column mean_weight, then summarize# A tibble: 3 × 2
sex mean_weight
<chr> <dbl>
1 F 42.2
2 M 43.0
3 <NA> 64.7Notice that we can pass summarize a column name – so it’s almost like saying “give me a tibble with this column that is this values/computational output”.
You can also group by multiple columns.
surveys %>%
group_by(sex, species_id) %>% # create more granular groups of sex, species_id combined
summarize(mean_weight = mean(weight, na.rm = TRUE)) %>%
tail()`summarise()` has grouped output by 'sex'. You can override using the `.groups`
argument.# A tibble: 6 × 3
# Groups: sex [1]
sex species_id mean_weight
<chr> <chr> <dbl>
1 <NA> SU NaN
2 <NA> UL NaN
3 <NA> UP NaN
4 <NA> UR NaN
5 <NA> US NaN
6 <NA> ZL NaNHere, we used tail() to look at the last six rows of our summary. Before, we had used head() to look at the first six rows.
We can see that the sex column contains NA values. The resulting mean_weight column does not contain NA but NaN (which refers to “Not a Number”) because mean() was called on a vector of NA values while at the same time setting na.rm = TRUE (we didn’t filter out the rows, we just omitted the NAs from the calculation).
To avoid this, we can remove the missing values for weight before we attempt to calculate the summary statistics on weight. Because the missing values are removed first, we can omit na.rm = TRUE when computing the mean.
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight))`summarise()` has grouped output by 'sex'. You can override using the `.groups`
argument.# A tibble: 64 × 3
# Groups: sex [3]
sex species_id mean_weight
<chr> <chr> <dbl>
1 F BA 9.16
2 F DM 41.6
3 F DO 48.5
4 F DS 118.
5 F NL 154.
6 F OL 31.1
7 F OT 24.8
8 F OX 21
9 F PB 30.2
10 F PE 22.8
# … with 54 more rowsOnce the data are grouped, you can also summarize multiple variables at the same time (and not necessarily on the same variable).
For instance, we could add a column indicating the minimum weight for each species for each sex.
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight))`summarise()` has grouped output by 'sex'. You can override using the `.groups`
argument.# A tibble: 64 × 4
# Groups: sex [3]
sex species_id mean_weight min_weight
<chr> <chr> <dbl> <dbl>
1 F BA 9.16 6
2 F DM 41.6 10
3 F DO 48.5 12
4 F DS 118. 45
5 F NL 154. 32
6 F OL 31.1 10
7 F OT 24.8 5
8 F OX 21 20
9 F PB 30.2 12
10 F PE 22.8 11
# … with 54 more rowsWe can also arrange the data.
For example, let’s sort (low to high value) on min_weight.
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight)) %>%
arrange(min_weight)`summarise()` has grouped output by 'sex'. You can override using the `.groups`
argument.# A tibble: 64 × 4
# Groups: sex [3]
sex species_id mean_weight min_weight
<chr> <chr> <dbl> <dbl>
1 F PF 7.97 4
2 F RM 11.1 4
3 M PF 7.89 4
4 M PP 17.2 4
5 M RM 10.1 4
6 <NA> PF 6 4
7 F OT 24.8 5
8 F PP 17.2 5
9 F BA 9.16 6
10 M BA 7.36 6
# … with 54 more rowsWe can sort in descending order by using the desc() function.
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight)) %>%
arrange(desc(mean_weight))`summarise()` has grouped output by 'sex'. You can override using the `.groups`
argument.# A tibble: 64 × 4
# Groups: sex [3]
sex species_id mean_weight min_weight
<chr> <chr> <dbl> <dbl>
1 <NA> NL 168. 83
2 M NL 166. 30
3 F NL 154. 32
4 M SS 130 130
5 <NA> SH 130 130
6 M DS 122. 12
7 <NA> DS 120 78
8 F DS 118. 45
9 F SH 78.8 30
10 F SF 69 46
# … with 54 more rowsCounting
We often want to count the number of observations found for each factor or a combination of factors. For this, we use count().
Let’s count the number of rows of data for each sex.
surveys %>%
count(sex)# A tibble: 3 × 2
sex n
<chr> <int>
1 F 15690
2 M 17348
3 <NA> 1748The count() function is shorthand for something we’ve already seen: grouping by a variable, and summarizing it by counting the number of observations in that group. In other words, surveys %>% count() is equivalent to
surveys %>%
group_by(sex) %>%
summarize(count = n())# A tibble: 3 × 2
sex count
<chr> <int>
1 F 15690
2 M 17348
3 <NA> 1748For convenience, count() provides the sort argument.
surveys %>%
count(sex, sort = TRUE)# A tibble: 3 × 2
sex n
<chr> <int>
1 M 17348
2 F 15690
3 <NA> 1748Previous example shows the use of count() to count the number of rows/observations for one factor (i.e., sex).
If we wanted to count combination of factors, such as sex and species, we would specify the first and the second factor as the arguments of count()
surveys %>%
count(sex, species)# A tibble: 81 × 3
sex species n
<chr> <chr> <int>
1 F albigula 675
2 F baileyi 1646
3 F eremicus 579
4 F flavus 757
5 F fulvescens 57
6 F fulviventer 17
7 F hispidus 99
8 F leucogaster 475
9 F leucopus 16
10 F maniculatus 382
# … with 71 more rowsWith the above code, we can then use arrange() to sort the table according to a number of criteria so that we have a better comparison.
For instance, we might want to arrange the table above in
- an alphabetical order of the levels of the species
- in descending order of the count
surveys %>%
count(sex, species) %>%
arrange(species, desc(n)) #n is the column name of the counts output from the count command, so it's used here to define the decreasing on value# A tibble: 81 × 3
sex species n
<chr> <chr> <int>
1 F albigula 675
2 M albigula 502
3 <NA> albigula 75
4 <NA> audubonii 75
5 F baileyi 1646
6 M baileyi 1216
7 <NA> baileyi 29
8 <NA> bilineata 303
9 <NA> brunneicapillus 50
10 <NA> chlorurus 39
# … with 71 more rowsFrom the table above, we may learn that, for instance, there are 75 observations of the albigula species that are not specified for its sex (i.e. NA).
Challenge
Q&A: Using survey, how many animals were caught in each plot_type surveyed?
Challenge
Q&A: Starting with survey, use group_by() and summarize() to find the mean, min, and max hindfoot length for each species (using species_id). Also add the number of observations (hint: see ?n).
Challenge
Q&A: Starting with survey, what was the heaviest animal measured in each year? Return the columns year, genus, species_id, and weight.
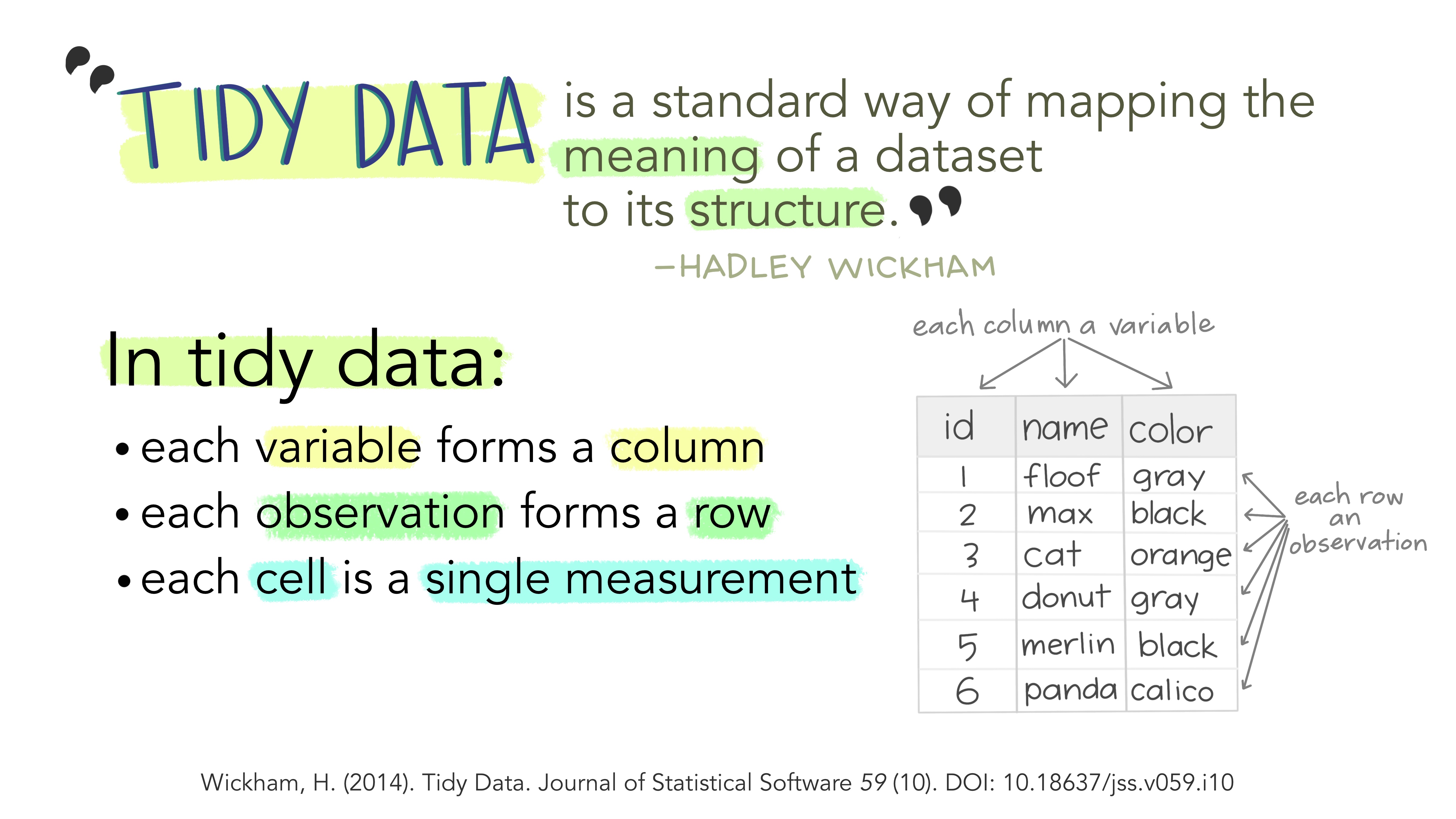
Tidy data
Structuring data is an important aspect of working with data, making it easier to analyze and visualize. The best way to work with our data is in “tidy” format.
Reshaping with pivot_longer() and pivot_wider()
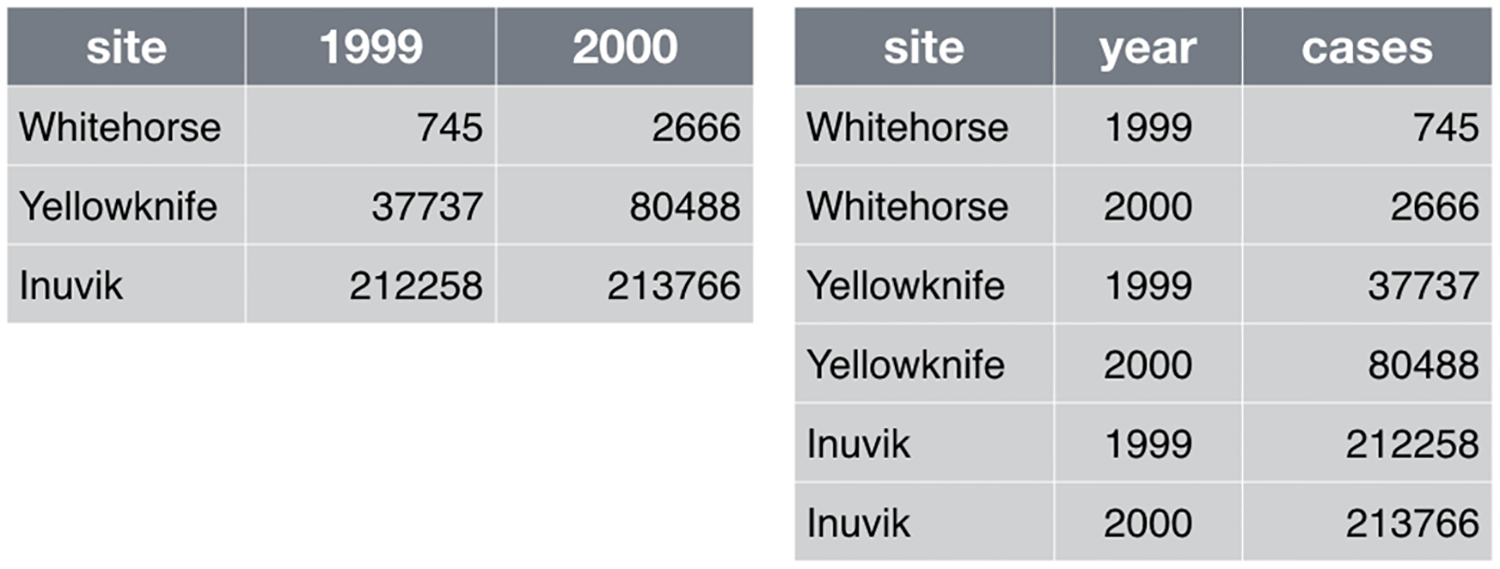
When working with tidy data there are two common dataformats for tabular datasets, wide and long.
In the example below we collected information on 3 variables – site, year, and cases.
Wide
- Data relating to the same measured thing in different columns.
- In this case, we have values related to our “metric” spread across multiple columns (a column each for a year).
Long
- A column for each of the types of things we measured or recorded in our data.
- In other words, each variable has its own column (a column for site, year, and case).
Depending on what you want do to, you might want to reshape from one form to the other. Luckily, there are functions for the pivot_wider() and pivot_longer().
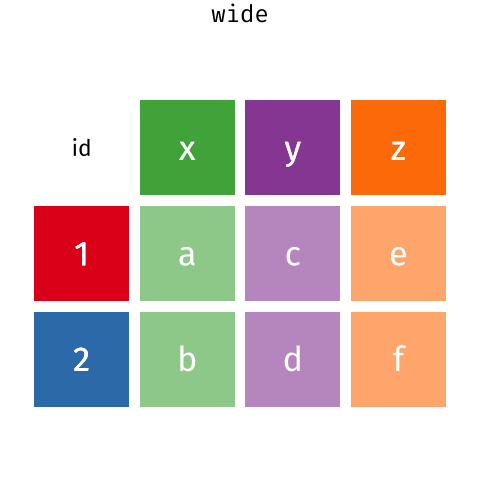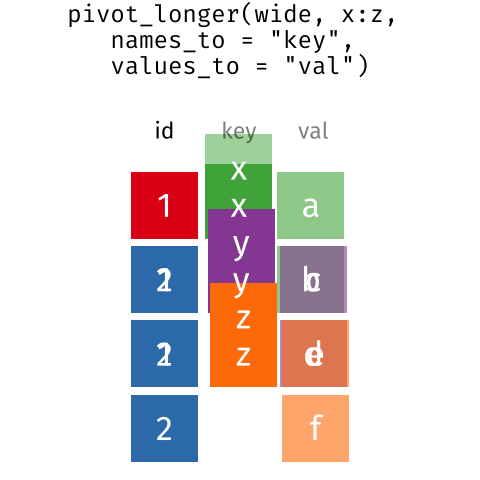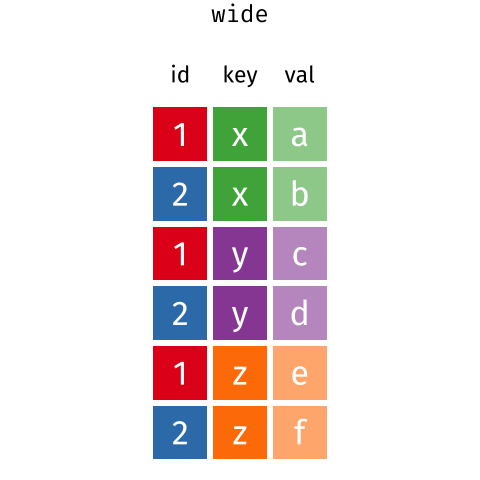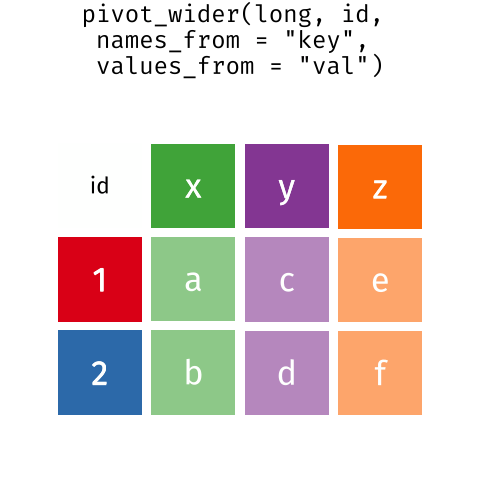
Pivoting from long to wide format
pivot_wider() takes three principal arguments
- the data
- the names_from column variable whose values will become new column names.
- the values_from column variable whose values will fill the new column variables.
Further arguments include values_fill which, if set, fills in missing values with the value provided.
What if we wanted to plot comparisons between the weight of genera in different plots?
To do this, we’ll need to 1. Create our dataset of interest – will generate a long formatted dataframe 2. Convert from long to wide format, filling missing values with 0.
Create our dataset – use filter(), group_by() and summarize() to filter our observations and variables of interest, and create a new variable for the mean_weight.
surveys_gw <- surveys %>%
filter(!is.na(weight)) %>% #remove rows with empty weight
group_by(plot_id, genus) %>% #group by plot_id and genus
summarize(mean_weight = mean(weight)) #give me the mean weight for each group`summarise()` has grouped output by 'plot_id'. You can override using the
`.groups` argument.head(surveys_gw)# A tibble: 6 × 3
# Groups: plot_id [1]
plot_id genus mean_weight
<dbl> <chr> <dbl>
1 1 Baiomys 7
2 1 Chaetodipus 22.2
3 1 Dipodomys 60.2
4 1 Neotoma 156.
5 1 Onychomys 27.7
6 1 Perognathus 9.62This yields surveys_gw, a long-formatted dataframe where the observations for each plot are distributed across multiple rows, 196 observations of 3 variables.
Let’s use pivot_wider() as follows
data– thesurveys_gwdataframenames_from–genus(column variable whose values will become new column names)values_from–mean_weight(column variable whose values will fill the new column variables)
Using pivot_wider() with the names from genus and with values from mean_weight this becomes 24 observations of 11 variables, one row for each plot.
surveys_wide <- surveys_gw %>%
pivot_wider(names_from = genus, values_from = mean_weight, values_fill = 0)
head(surveys_wide)# A tibble: 6 × 11
# Groups: plot_id [6]
plot_id Baiomys Chaetodipus Dipodomys Neotoma Onycho…¹ Perog…² Perom…³ Reith…⁴
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 7 22.2 60.2 156. 27.7 9.62 22.2 11.4
2 2 6 25.1 55.7 169. 26.9 6.95 22.3 10.7
3 3 8.61 24.6 52.0 158. 26.0 7.51 21.4 10.5
4 4 0 23.0 57.5 164. 28.1 7.82 22.6 10.3
5 5 7.75 18.0 51.1 190. 27.0 8.66 21.2 11.2
6 6 0 24.9 58.6 180. 25.9 7.81 21.8 10.7
# … with 2 more variables: Sigmodon <dbl>, Spermophilus <dbl>, and abbreviated
# variable names ¹Onychomys, ²Perognathus, ³Peromyscus, ⁴Reithrodontomys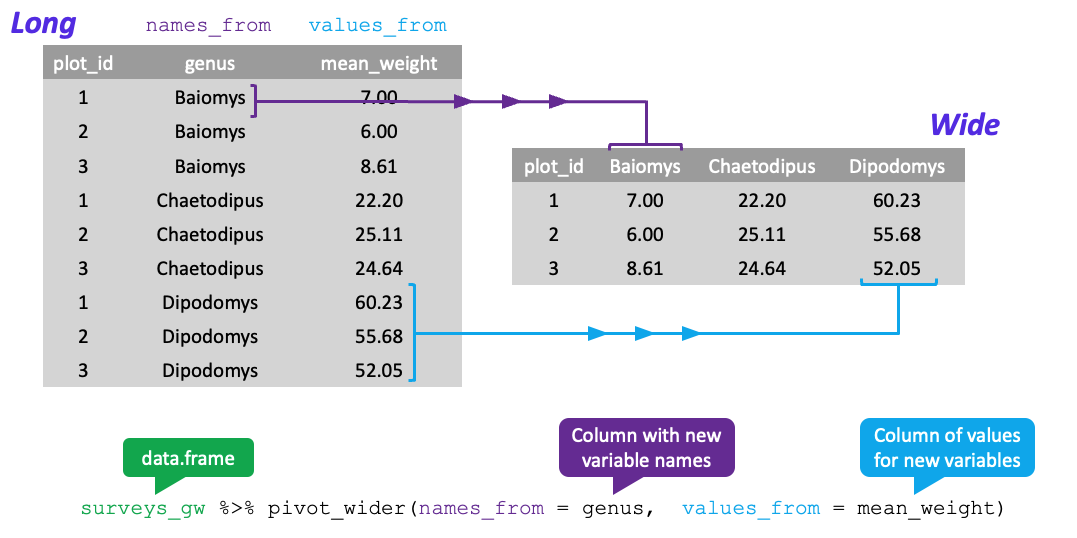
Pivoting from wide to long format
What if we wanted were given a wide format, but wanted to treat the genus names (which are column names) as values of a genus variable instead (we wanted a genus column)?
pivot_longer() takes four principal arguments:
- the data
- the names_to column variable we wish to create from column names.
- the values_to column variable we wish to create and fill with values.
- cols are the name of the columns we use to make this pivot (or to drop).
Let’s use pivot_longer() as follows
data– thesurveys_widedataframenames_to–genus(column variable to create from column names)values_to–mean_weight(column variable we wish to create and fill with values)cols–-plot_id(names of columns we use to make the pivot or drop)
surveys_long <- surveys_wide %>%
pivot_longer(names_to = "genus", values_to = "mean_weight", cols = -plot_id) #were including all cols here except plot_id
head(surveys_long)# A tibble: 6 × 3
# Groups: plot_id [1]
plot_id genus mean_weight
<dbl> <chr> <dbl>
1 1 Baiomys 7
2 1 Chaetodipus 22.2
3 1 Dipodomys 60.2
4 1 Neotoma 156.
5 1 Onychomys 27.7
6 1 Perognathus 9.62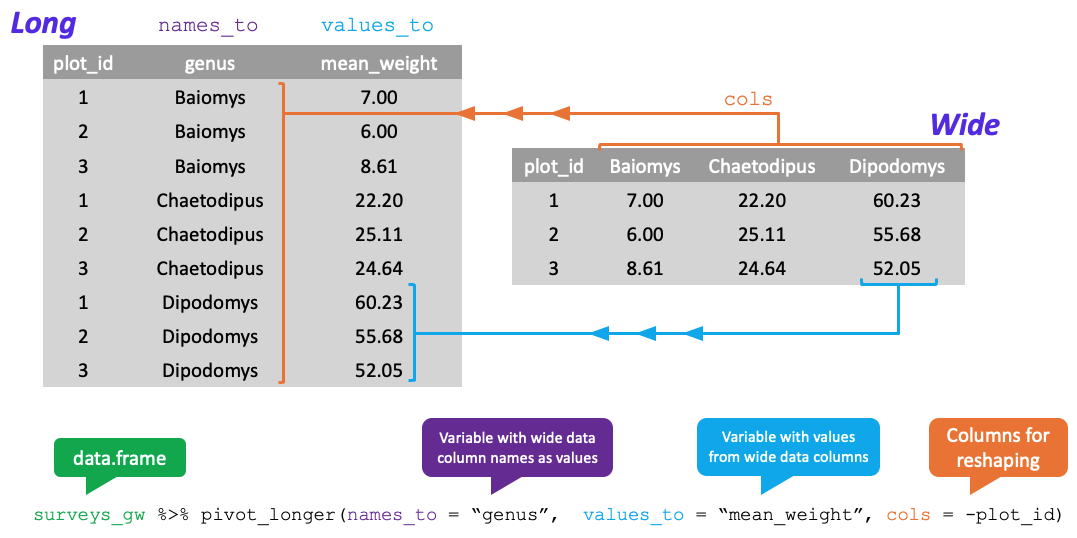
Challenge
Q&A: Reshape the surveys dataframe with year as columns, plot_id as rows, and the number of genera per plot as the values.
You will need to summarize before reshaping, and use the function n_distinct() to get the number of unique genera within a particular chunk of data. It’s a powerful function! See ?n_distinct for more.
Challenge
Q&A: Now take that data frame and pivot_longer() it, so each row is a unique plot_id by year combination.
Exporting data
After analyzing, processing data, we often want to export the results to a file.
Let’s create a cleaned up version of our dataset that does not include missing data (weight, hindfoot_length, and sex) – in preparation for next session.
surveys_complete <- surveys %>%
filter(!is.na(weight), # remove missing weight
!is.na(hindfoot_length), # remove missing hindfoot_length
!is.na(sex)) # remove missing sexWe’re going to be plotting species abundances over time – so let’s remove observations for rare species (that have been observed less than 50 times).
## Extract the common species_id
species_counts <- surveys_complete %>%
count(species_id) %>%
filter(n >= 50)
## Use the common species id as a key for filtering (keeping)
surveys_complete <- surveys_complete %>%
filter(species_id %in% species_counts$species_id) To make sure that everyone has the same dataset, check the dimensions by typing dim(surveys_complete), with values matching the output shown.
dim(surveys_complete)[1] 30463 13Now, let’s write to a file with write_csv().
write_csv(surveys_complete, file = "~/data-carpentry/data/surveys_complete.csv")Citations
- Data Analysis and Visualization in R for Ecologists. https://datacarpentry.org/R-ecology-lesson/index.html
- Wickham, H. Tidy Data. Journal of Statistical Software 59 (10). https://10.18637/jss.v059.i10